-Search query
-Search result
Showing 1 - 50 of 58 items for (author: weber & j)

EMDB-19562: 
Vimentin intermediate filament protofibril stoichiometry
Method: subtomogram averaging / : Eibauer M, Medalia O

EMDB-19563: 
Vimentin intermediate filament structure (delta tail)
Method: helical / : Eibauer M, Medalia O

EMDB-16844: 
Vimentin intermediate filament structure
Method: helical / : Eibauer M, Medalia O

EMDB-16024: 
SARS-CoV-2 S protein in complex with pT1644 Fab
Method: single particle / : Stroeh L, Hansen G, Vollmer B, Krey T, Benecke T, Gruenewald K

EMDB-16026: 
SARS-CoV-2 S protein in complex with pT1696 Fab
Method: single particle / : Hansen G, Benecke T, Vollmer B, Gruenewald K, Krey T

EMDB-28205: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 2
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28206: 
Clostridioides difficile binary toxin translocase CDTb wild-type after calcium depletion from receptor binding domain 1 (RBD1) - Class 1
Method: single particle / : Abeyawardhane DL, Pozharski E

EMDB-28207: 
Clostridioides difficile binary toxin translocase CDTb double mutant - D623A D734A
Method: single particle / : Abeyawardhane DL, Pozharski E
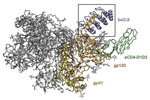
EMDB-26157: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J

PDB-7txd: 
Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9
Method: single particle / : Cerutti G, Gorman J, Kwong PD, Shapiro L

EMDB-15987: 
In situ sub-tomogram average of the Ca. L. ossiferum ribosome
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15988: 
In situ sub-tomogram average of Ca. L. ossiferum actin filament
Method: subtomogram averaging / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15989: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15990: 
cryo-tomogram of Ca. L. ossiferum protrusions
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15991: 
cryo-tomogram of a Ca.L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15992: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-15993: 
cryo-tomogram of a Ca. L. ossiferum cell
Method: electron tomography / : Rodrigues-Oliveira T, Wollweber F, Ponce-Toledo R, Xu J, Rittmann S, Klingl A, Pilhofer M, Schleper C

EMDB-13694: 
Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state A)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

EMDB-13695: 
Open-gate mycobacterium 20S CP proteasome in complex MPA - global 3D refinement
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

EMDB-13696: 
Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state B)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

EMDB-13697: 
Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

EMDB-13698: 
Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state B)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

PDB-7px9: 
Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state A)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

PDB-7pxa: 
Open-gate mycobacterium 20S CP proteasome in complex MPA - global 3D refinement
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

PDB-7pxb: 
Substrate-engaged mycobacterial Proteasome-associated ATPase - focused 3D refinement (state B)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

PDB-7pxc: 
Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

PDB-7pxd: 
Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state B)
Method: single particle / : Jomaa A, Kavalchuk M, Weber-Ban E

EMDB-11002: 
CryoEM structure of bovine cytochrome bc1 in complex with a tetrahydroquinolone inhibitor
Method: single particle / : Muench SP, Johnson R, Amporndanai K, Atonyuk S

EMDB-10259: 
Sub-tomogram average of the Nap adhesion complex from the human pathogen Mycoplasma genitalium.
Method: subtomogram averaging / : Scheffer MP, Aparicio D

EMDB-10260: 
Structure the Nap adhesion complex from the human pathogen Mycoplasma genitalium by single particle cryo-EM.
Method: single particle / : Scheffer MP, Aparicio D

EMDB-10890: 
cryo-electron density map of the P140-P110 heterodimer
Method: single particle / : Scheffer MP, Aparicio D

PDB-6yrk: 
P140-P110 complex fitted into the cryo-electron density map of the heterodimer
Method: single particle / : Scheffer MP, Aparicio D

EMDB-10319: 
Cryo-EM structure of the full-length ABC-transporter IrtAB
Method: single particle / : Weber MS, Arnold F, Gonda I, Seeger M, Medalia O

EMDB-20926: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation
Method: single particle / : Xu X, Pozharski E

EMDB-20927: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation
Method: single particle / : Xu X, Pozharski E

PDB-6uwr: 
Clostridium difficile binary toxin translocase CDTb in asymmetric tetradecamer conformation
Method: single particle / : Xu X, Pozharski E, des Georges A

PDB-6uwt: 
Clostridium difficile binary toxin translocase CDTb tetradecamer in symmetric conformation
Method: single particle / : Xu X, Pozharski E, des Georges A

EMDB-21001: 
Cryo-EM structure of porcine ATP synthase reconstituted in small unilamellar vesicles_I
Method: single particle / : Mnatsakanyan N, Llaguno MC, Yang Y, Yan Y, Weber J, Sigworth FJ, Jonas EA

EMDB-21002: 
Cryo-EM structure of porcine ATP synthase reconstituted in small unilamellar vesicles_II
Method: single particle / : Mnatsakanyan N, Llaguno MC, Yang Y, Yan Y, Weber J, Sigworth FJ, Jonas EA

EMDB-4584: 
Structure and assembly of the mitochondrial membrane remodelling GTPase Mgm1
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel J, Wollweber F, Pfitzner A, Muehleip A, Sanchez R, Kudryashev M, Chiaruttin N, Lilie H, Schleger J, Rosenbaum E, Hessenberger M, Matthaeus C, Noe F, Roux A, van der Laan M, Kuehlbrandt W, Daumke O
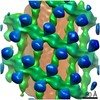
EMDB-10062: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O
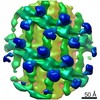
EMDB-10063: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10064: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-10065: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzt: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuehlbrandt W, Daumke O

PDB-6rzu: 
Structure of s-Mgm1 decorating the outer surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzv: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

PDB-6rzw: 
Structure of s-Mgm1 decorating the inner surface of tubulated lipid membranes in the GTPgammaS bound state
Method: subtomogram averaging / : Faelber K, Dietrich L, Noel JK, Sanchez R, Kudryashev M, Kuelbrandt W, Daumke O

EMDB-3561: 
map of the RNA polymerase lambda-based antitermination complex solved by cryo-EM
Method: single particle / : Said N, Krupp F
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
